GC-Rich Cloning of HLA Nonclassical Class I Heavy Chain
PCR Success Story #21Project Description
Story #21: GC-Rich Cloning HLA Nonclassical Class I Heavy Chain (paid service).
A Montreal researcher contacted us to perform the cloning of a Human Leukocyte Antigen (HLA) Heavy Chain from human genomic DNA. Firstly, in order to perform GC-Rich Cloning of their genomic sequence of interest, High-Fidelity PCR had to be optimized. Secondly, after gel extraction of the PCR product, cloning had to be performed using a topoisomerase blunt cloning kit. Subsequently, high-fidelity PCR had to be performed by colony PCR. Finally, the researcher himself would take care of DNA sequencing afterwards.
To protect the research and privacy of the customer who paid for our Cloning Services, the exact name of the gene of interest and researcher are held confidential.
GC-Rich HLA DNA Sequence
Target lenght: 1717 bp
Average GC content over 1717 bp: 62 %
Average GC content of bases 17 to 553 containing 6 GC-rich segments : 70%
- GC-rich segment 1 spans over 37 bp with 92% GC
- GC-rich segment 2 spans over 56 bp with 79% GC
- GC-rich segment 3 spans over 90 bp with 77% GC
- GC-rich segment 4 spans over 90 bp with 77% GC
- GC-rich segment 5 spans over 67 bp with 84% GC
- GC-rich segment 6 spans over 39 bp with 79% GC
Project Details
Client: CR-CHUM
Date: January 30th, 2017
Type of experiment: GC-Rich PCR, GC-rich PCR, Blunt Cloning, Colony PCR
DNA Polymerase: TransStart FastPfu FLY (Ultra-HiFi) DNA Polymerase
Competitor: Q5® High-Fidelity DNA Polymerase. Q5® is a trademark of New England Biolabs, Inc.
PCR Optimization for GC-rich Cloning of the HLA gene
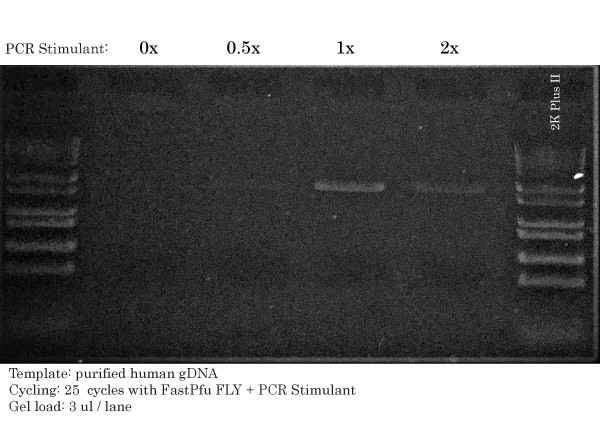
High-Fidelity PCR performed with FastPfu FLY (Ultra-HiFi) DNA Polymerase. A gradient of PCR Stimulant was used to loosen the gDNA and bring down its melting temperature. PCR was optimal between a concentration of 1X and 2X of PCR Stimulant.
PCR setup for High-Fidelity GC-rich PCR:
- H2O : to 20 ul
- 5x buffer : 4 ul
- PCR Stimulant (5x): 2, 4 or 8 ul
- dNTPs (2.5mM): 1.6 ul (0.2 mM final)
- F1 (10 uM): 0.4 ul (200 nM final)
- F2 (10 uM): 0.4 ul
- gDNA : 0.2 ul
- FastPfu FLY (2.5 u/ul) : 0.4 ul
PCR cycling for High-Fidelity GC-rich PCR
- Denaturation: 90s at 95 °C
- 25 x
- Denaturation: 15s at 95 °C
- Annealing: 20s at 50 °C
- Extension: 30s at 68 °C
- Final extension: 300s at 68 °C
Gel Extraction and GC-Rich Cloning by Topoisomerase
Prior to GC-Rich Cloning, the 1.7 kb amplicon was gel extracted using the FavorPrep GEL/PCR Purification Kit.
The blunt-ended purified DNA fragment was cloned with the pEASY-Blunt Zero Cloning kit. Then, 2 ul of the Topoisomerase-mediated cloning reaction was transformed into 50 ul of Trans1-T1 Phage Resistant Chemically Competent Cells. Subsequently, cells were plated on LB-agar Ampicillin plates.
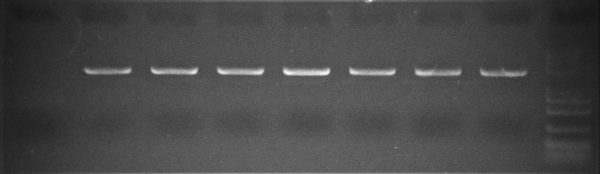
Colony PCR after GC-rich Cloning
Colonies were picked and DNA extracted with TransDirect Animal Tissue Buffer Set. Then, High-Fidelity PCR was performed with 2x TransStart® FastPfu PCR SuperMix without any addition of PCR Stimulant, The same PCR Cycling protocol was used as with FastPfu FLY.
Sequencing
PCR Products were sequenced by the researcher and they found allelic mutants of interest 🙂
Now Here Comes the Fun Part!
The Comparison of FastPfu FLY with Q5 High-Fidelity DNA PolymeraseWant us to Perform and Design your Cloning Project too?
Put us in charge !